The progressive shift towards an older demographic creates an urgent need for innovative and multidisciplinary research to predict soft tissue changes throughout the entire organ life-cycle. There is tremendous scientific opportunity for engineering disciplines to contribute to our understanding of living matter and its continuous adaptation of anatomical structure and mechanobiochemical properties on the cellular, tissue, and organ level. Our ultimate goal is to utilize combined computational and experimental methods to advance translational research in technologically and scientifically challenging problems of living matter physics.
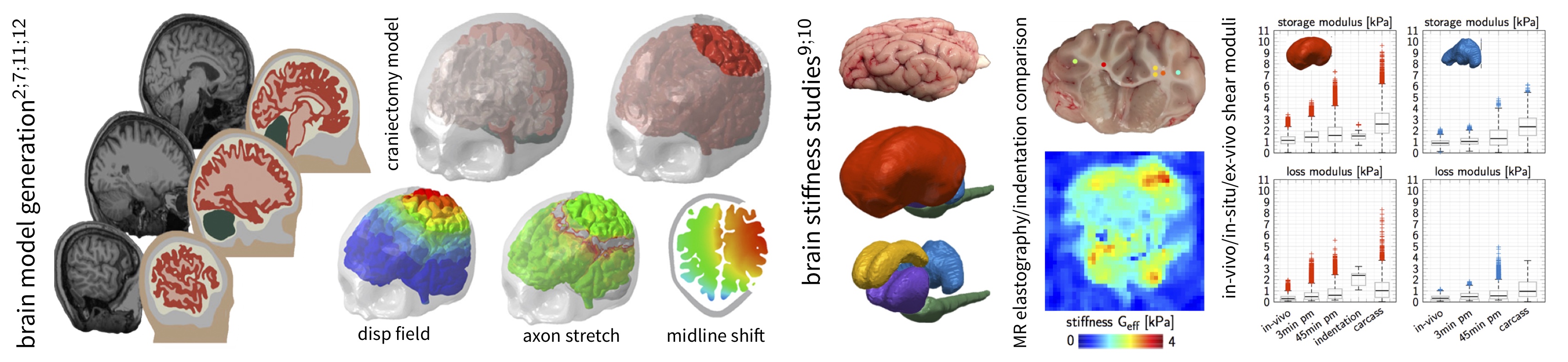
Personalized brain model for simulations of a decompressive craniectomy to assess axonal loading; In-vivo, in-situ, and ex-vivo brain stiffness measurements using magnetic resonance elastography and indentation. A brain specific segmentation atlas enables an automatic analysis of cortical substructures including cerebrum, cerebellum, corpus callosum, thalamus, and brainstem. MRE based dynamic moduli shown for the cerebrum and cerebellum under in-vivo, in-situ, and ex-vivo conditions.
Our Expertise
Material Modeling and Finite Element Simulations
Skin, muscle, and the brain are characterized by a highly nonlinear, viscoelastic, and anisotropic tissues behavior and generally respond to environmental factors or disease by comprehensive morphological and biochemical adaptations across temporal and spatial scales. Tissue specific constitutive equations describing these mechanical phenomena require a good understanding of the tissue’s constituents individual contributions ranging from the cell to the organ level. We develop and implement constitutive material models and finite element formulations for medically relevant numerical simulations. We use a generalized multi-field finite element approach to couple the deformation problem to biochemical diffusion and transport equations in order to capture the biophysical response of muscle, skin, and the brain.
Our simulations are based on anatomically accurate, numerically reliable, and predictive finite element models of organs. Based on semi-automatic medical image segmentation and meshing, we work with several subject-specific computational models, including a brain model (in collaboration with Simpleware), a highly detailed forehead model (in collaboration with the University Hospital Zurich), a multi-layer face model (in collaboration with Johnson&Johnson), and a mastication model (in collaboration with the Nestlé Research Center).
Experimental Characterization of Soft Tissues
Mechanical modeling of soft biological tissues heavily relies on an experimental visualization and quantification of stress, strain, and biochemical changes. Newly developed in-vivo measurement methods hold the key to incorporate mechanical parameters in translational and patient-specific medical care. Our lab specializes in indentation testing of soft biological tissues to determine the nonlinear and time dependent material response to inform our constitutive models. Our expertise also extends to the mechanical characterization of facial skin tissue using suction-based multi-axial loading methods, as well as medical imaging and ultrasonography to visualize the deformation mechanisms of muscle and skin. We also collaborate with other research labs (Kurt Lab at Stevens, Center for Biomedical Imaging at Stanford) for in-vivo and in-situ brain stiffness measurements using magnetic resonance elastography.
Our experimental work is driven by the necessity to understand soft tissue behavior across temporal and spatial scales in order to inform finite element simulations. The predictive capabilities of our computational tools are validated through the direct comparison of experimentally observed and numerically calculated tissue behavior.
Current Research Interests of our Lab
Life Cycle Analysis of the Human Brain
Numerous research initiatives have brought us closer than ever to understanding the molecular mechanisms of neurodegeneration; yet, to date, there is no unified, integrative technology to characterize and predict the progression of neurodegeneration in space and time. Our long-term goal is to determine a prognostic, physiologically mechanistic tool to simulate the life cycle of the human brain and predict how its physiological form, function, and behavior change under pathological conditions. While the concise etiology of Alzheimer’s disease, dementia, chronic traumatic encephalopathy, and multiple sclerosis, is still poorly understood, it is becoming evident that the localized activation of microglia through normal brain aging or diffuse axonal injury is central to the initiation of a neuroinflammatory response and the spread of abnormal proteins. We use a multi-scale, multi-field approach to connect the relevant scales in the living brain and to inform continuum field theories of characteristic biophysical features such as brain aging, toxic protein aggregation, disease propagation, and neurodegeneration.
The Role of Myelin in Neuro-Mechanobiology
Myelinogenesis, or the wrapping of myelin sheaths around axon fibers, is one of the most important post-natal mechanobiological changes in the brain. Axon myelination by oligodendrocytes, which are part of the understudied microglial cell network building the scaffold of the brain, occurs during the first two decades in life and represents the brain’s exhaustive adaptation to increase signaling speed between functionally related regions. Normal brain aging, and demyelinating diseases like multiple sclerosis in particular, lead to a significant loss of white matter tissue and behavioral changes associated with a decline in memory, language, and other cognitive skills. The combination of medical imaging and clinically validated computational tools will provide fundamentally new perspectives on personalized treatment methods of myelin-related diseases.
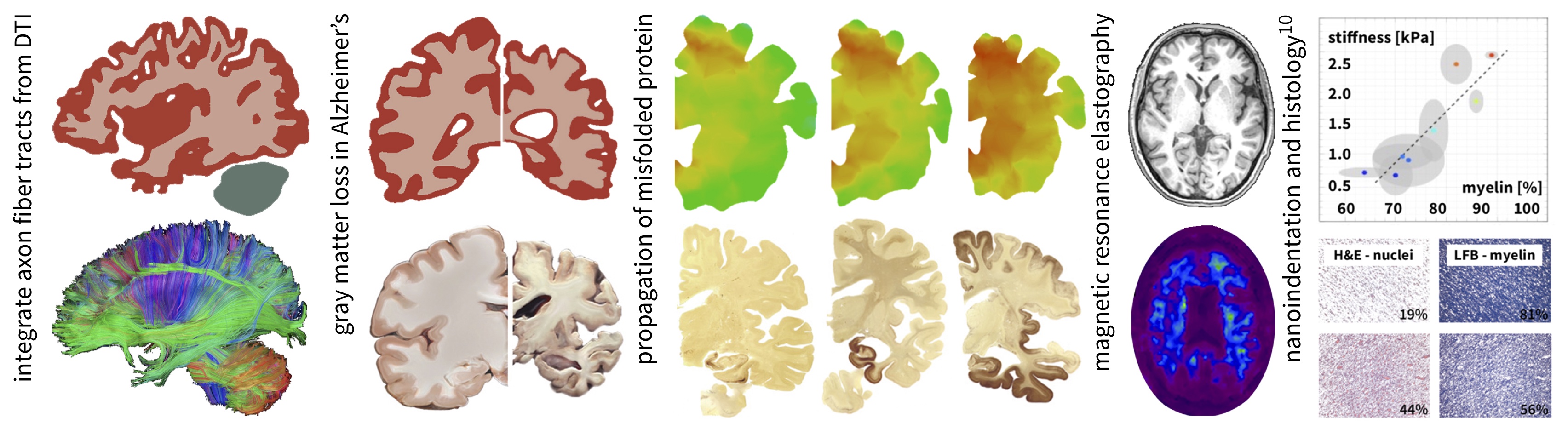
Axon fiber tracts from diffusion tensor imaging; simulations of neurodegeneration and disease propagation in the brain; investigating the role of myelin in development and disease.